- ABI - SOFT group
Missions
ABI-Soft team aims to develop, deploy and provide analysis tools and databases useful for scientists of the lab. The team also provides support to development projects carried out by the research teams of the lab.
Abi-Soft aims also to make them available to the plant science community through the French national forge for research and higher education (SourceSup or forgemia) and efficient deployment solutions (virtual machines, containers).
The team is also in charge of the training of the scientists (local and their partners) to use the tools developed.
ABI-Soft works in close collaboration with the ABI-SYS team which maintains the infrastructure hosting the local instances of our applications.
Currently, ABI-Soft develops several data management tools (Thaliadb, SHiNeMaS and DiverCILand) and analysis tools (BioMercator and OptimaRs) that are components of a global information system dedicated to plant breeding.
Current projects
ThaliaDB
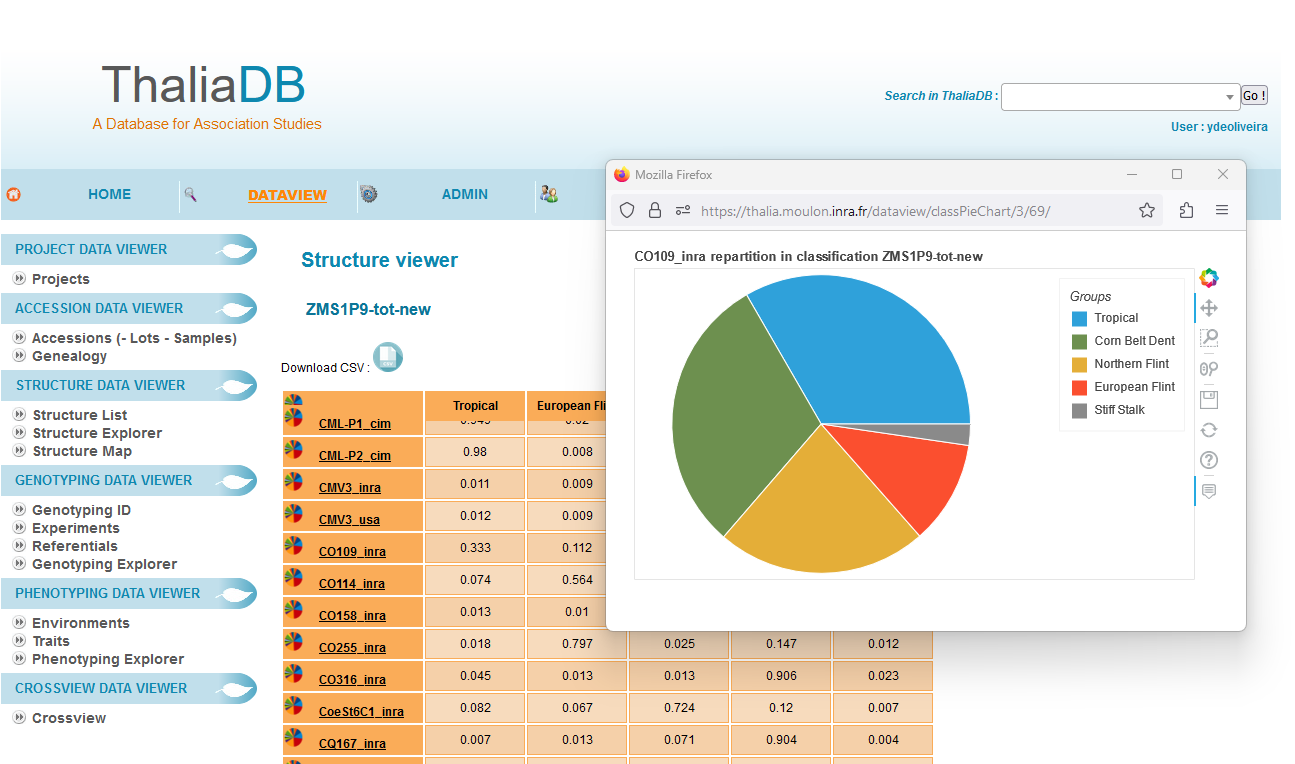
In collaboration with GQMS team. ThaliaDB is a web application with its database that manages plant accessions, seeds lots, SNP markers, genotyping rawddata and field phenotyping elaborated data.
This tool is useful for lab data management, for data curation, to prepare datasets for genome wide assocation studies or genomic selection analysis and for data sharing with partners and external data repositories (Dataverse). ThaliaDB screen
SHiNeMaS
SHiNeMaS screen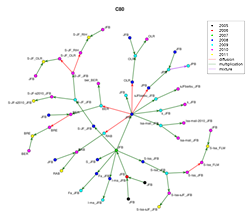
DiverCILand
DiverCILand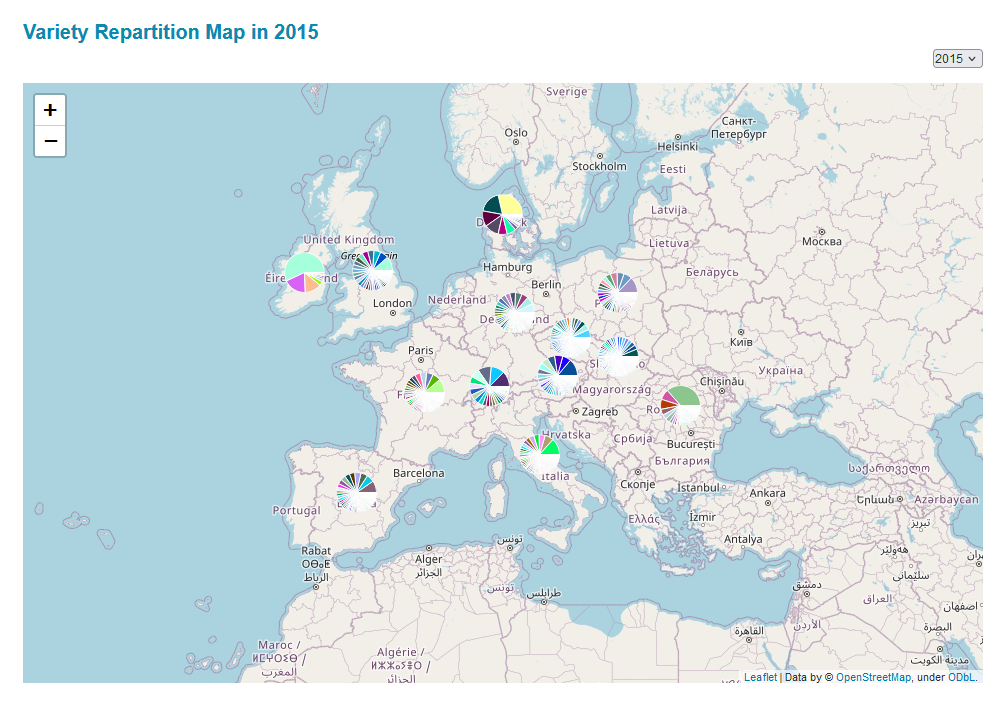
Biomercator
In collaboration with
GQMS
team.
Biomercator screen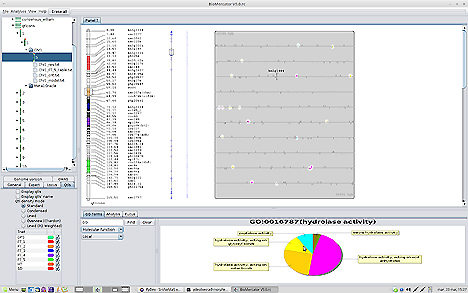
The new version currently in development (V5) will provide tools to explore GWAS analyses.
OptimaRs
OptimaRs is a decision support tool to conduct Marker-Assisted Selection programmes.
OptimaRs screen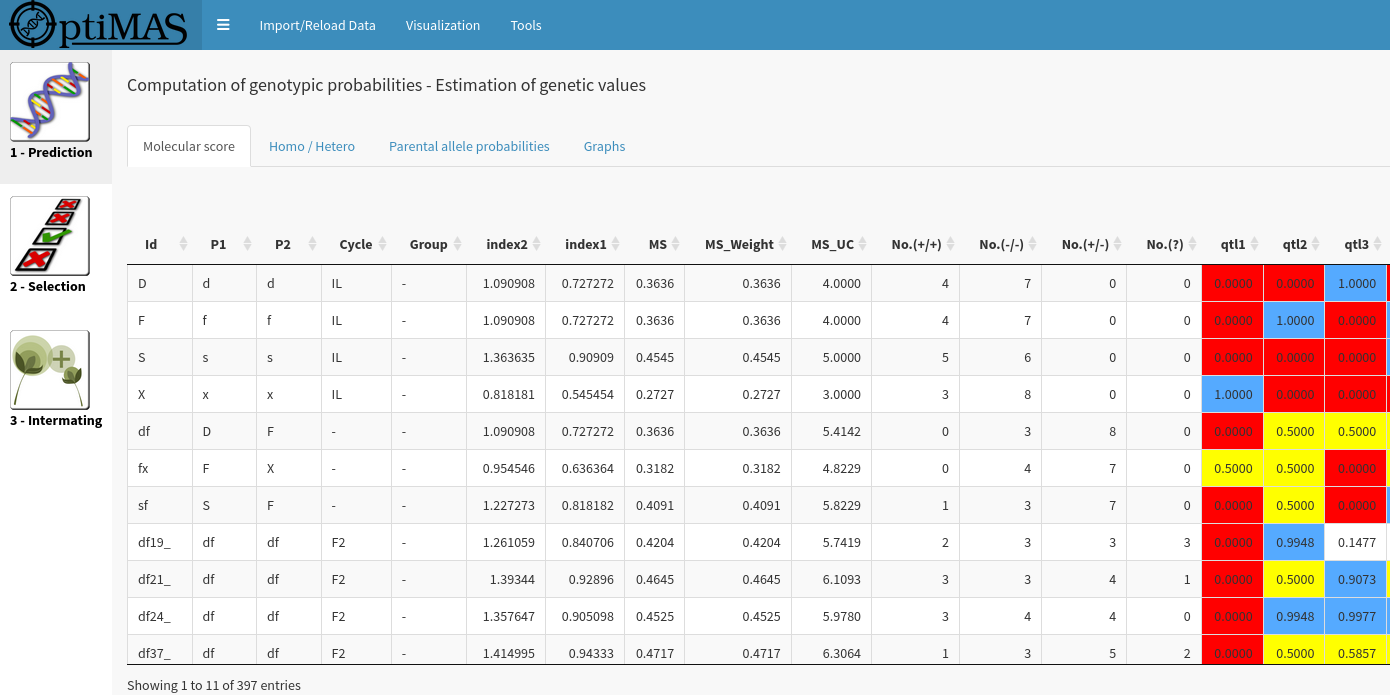
OptimaRs includes in a Graphical User Interface (GUI), three different modules corresponding to the different steps of a selection programme :
Using information given by markers located in the vicinity of the estimated QTL positions, probabilities of favorable parental QTL allele transmission are computed in different MAS schemes and mating designs (intercrossing, selfing, backcrossing, double haploids, RIL) with the possibility of considering generations without genotypic information (step 1). Then strategies are proposed to select the best plants (step 2), and efficiently intermate them based on the expected value of their progenies (step 3).
Link to the project (version C++) : ![]()
Please contact Yannick De Oliveira, for more details and partnership on any project.
Previous staff
- Nesrine Mouhoubi (2020-2021), Master Data Science Paris-Saclay University
- Laetitia Courgey (2018-2020), Master MIAGE Paris Dauphine University (SHiNeMaS, ThaliaDB)
- Alice Beaugrand (2016-2018), Master 2 University Paris-Saclay, then CDD (SHiNeMaS, ThaliaDB)
- Artur Robieux (2018), Master 2 University Paris-Saclay (ThaliaDB)
- Marine Daniel-Dit Andrieu (2017), Master 2 University Paris-Saclay (Phenofield-Validator)
- Mélanie Polart-Donat (2017), Master 2 University Paris-Saclay (SHiNeMaS)
- Eva Dechaux (2017), Master 2 University Paris-Saclay (SHiNeMaS)
- Lan-Anh Nguyen (2016), DUT Université d’Auvergne, (ThaliaDB)
- Olivier Akmansoy (2016), Ecoles des Mines Douai (ThaliaDB)
- Aichatou Aboubacar-Amadou (2016), Master 2 Univ. Reims (SHiNeMaS)
- Lydia Aït-Brahim (2014 – 2016), Master 2 Université Paris-Saclay, then CDD (Biomercator)
- Guy Ross Assoumou (2012 – 2015), CDD IE/Software engineer (ThaliaDB)
- Jocelyn Chêne (2015), Master 2 Univ. Nantes (ANR Bakery project)
- Laura Burlot (2014), Master 2 Univ. Lyon, then CDD IE/ software engineer (SHiNeMaS)
- Marie Lefebvre (2014), Master2 Univ. Nantes (SHiNeMaS)
- Darkawi Madi (2012), Master 2 (SHiNeMaS)
Members
- Yannick De Oliveira Engineer (INRAE)
- Franck Gauthier Engineer (INRAE)
- Pierre Montalent Engineer (INRAE)
- Elise Peluso Licence student (INRAE)
- Mélanie Polart-Donat Engineer (INRAE)
Former members
Delphine Steinbach Research Engineer (1970), Laetitia Courgey Master Apprentissage (2018-2020)Publications
- Vidal A., Gauthier F. , Rodrigez W., Guiglielmoni N., Leroux D., Chevrolier N., Jasson S., Tourrette E., Martin OC., Falque M.. (2022) SeSAM: software for automatic construction of order-robust linkage maps. BMC Bioinformatics, 1 (23) 499
- De Oliveira Y. , Burlot L., Dawson JC., Goldringer I., Madi D., Rivière P., Steinbach D., van Frank G., Thomas M.. (2020) SHiNeMaS: a web tool dedicated to seed lots history, phenotyping and cultural practices. Plant Methods, 1 (16) 98
- Lopez Arias DC., Chastellier A., Thouroude T., Bradeen J., Van Eck L., De Oliveira Y. , Paillard S., Foucher F., Hibrand-Saint Oyant L., Soufflet-Freslon V.. (2020) Characterization of black spot resistance in diploid roses with QTL detection, meta-analysis and candidate-gene identification. Theor Appl Genet,
- Urien C., Legrand J., Montalent P. , Casaregola S., Sicard D.. (2019) Fungal Species Diversity in French Bread Sourdoughs Made of Organic Wheat Flour. Front. Microbiol., (10) 201
- Alaux M., Rogers J., Letellier T., Flores R., Alfama F., Pommier C., Mohellibi N., Durand S., Kimmel E., Michotey C., Guerche C., Loaec M., Lainé M., Steinbach D., Choulet F., Rimbert H., Leroy P., Guilhot N., Salse J., Feuillet C., Paux E., Eversole K., Adam-Blondon AF., Quesneville H., International Wheat Genome Sequencing Consortium. (2018) Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol, 1 (19) 111
- Adam-Blondon AF., Alaux M., Durand S., Letellier T., Merceron G., Mohellibi N., Pommier C., Steinbach D., Alfama F., Amselem J., Charruaud D., Choisne N., Flores R., Guerche C., Jamilloux V., Kimmel E., Lapalu N., Loaec M., Michotey C., Quesneville H., van Dijk ADJ.. (2017) Mining Plant Genomic and Genetic Data Using the GnpIS Information System. , (1533) 103-117
- Perronne R., Makowski D., Goffaux R., Montalent P. , Goldringer I.. (2017) Temporal evolution of varietal, spatial and genetic diversity of bread wheat between 1980 and 2006 strongly depends upon agricultural regions in France. Agriculture, Ecosystems & Environment, (236) 12-20
