
Natalia CONDE E SILVA
Genetics, Epigenetics and Evolution of Floral Morphogenesis
- Genetic
- Protein/Protein/DNA Interactions
- Plant biology
- Evolution
Université Paris-Saclay, MCF
natalia.conde-e-silva@universite-paris-saclay.fr — 01 69 15 34 11 — 1216
Génétique, Évolution et Écologie de la Morphologie Florale
- Génétique Quantitative et Évolution - Le Moulon
- Université Paris-Saclay, INRAE, CNRS, AgroParisTech
- IDEEV
- 12 route 128
- 91190 Gif-sur-Yvette
Position and Education
- 2015- : Associate Professor, UMR “Génétique Quantitative et Evolution – Le Moulon” (Université Paris Saclay, Gif-sur-Yvette, France).
- 2009-2015: Associate Professor, UMR “Institut de Biologie des Plantes” (Université Paris Sud, Gif-sur-Yvette, France).
- 2007-2009: ATER, Institut Jacques Monod (Université Paris-Diderot, Paris, France).
- 2007: PhD at Ecole doctorale “Inter///Bio” (Université Pierre et Marie Curie, Paris, France).
Biography and Research interests
Research communities and Societies
- RanOmics
- LabEx SPS (Sciences des Plantes de Saclay)
- GE2MorF is attached to the to SEVE doctoral school.
Teaching Activities
- Licence (Undergraduate): L2SDV : Biochimie (EN11048) ; Physiologie végétale (EN11053) ; L3SEM : Sciences et Jeunesse (DLSC304A) ; L3BS : Biochimie structurale et fonctionnelle (EN11096) ; L3BOE : Germination: de l’Organisme à la Molécule (EN11137) , TP de Biologie Moléculaire et Biochimie (EN11138) , Écophysiologie végétale (EN11135)
- Licence Double Diplôme : LDD1-MSV: Méthodologie (EN9715) ; LDD3-MSV/CSV Compléments en biologie moléculaire et biochimie (EN10768)
- Master (Graduate) : M1BIP : Adopte un gène (EN3612)
Research management
- 2020- : elected member to the scientific council of IDEEV
- 2020- : external member to the CSS “Biologie Intégrative des Plantes” (INRAE)
Publications
- Becker A., Bachelier JB., Carrive L., Conde e Silva N. , Damerval C., Del Rio C., Deveaux Y., Di Stilio VS., Gong Y., Jabbour F., Kramer EM., Nadot S., Pabón-Mora N., Wang W., RanOmics group. (2023) A cornucopia of diversity - Ranunculales as a model lineage. J Exp Bot, erad492
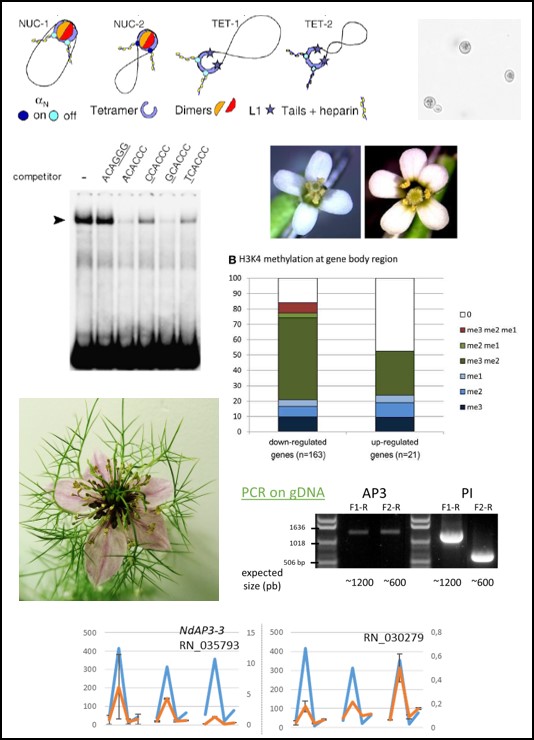
- Conde e Silva N. , Leguilloux M., Bellec A., Rodde N., Aubert J., Manicacci D., Damerval C., Berges H., Deveaux Y.. (2023) A MITE insertion abolishes the AP3-3 self-maintenance regulatory loop in apetalous flowers of Nigella damascena. Journal of Experimental Botany, 5 (74) 1448-1459
- Damerval C., Claudot C., Le Guilloux M., Conde e Silva N. , Brunaud V., Soubigou-Taconnat L., Caius J., Delannoy E., Nadot S., Jabbour F., Deveaux Y.. (2022) Evolutionary analyses and expression patterns of TCP genes in Ranunculales. Front Plant Sci, (13) 1055196
- Deveaux Y., Conde e Silva N. , Manicacci D., Le Guilloux M., Brunaud V., Belcram H., Joets J., Soubigou-Taconnat L., Delannoy E., Corti H., Balzergue S., Caius J., Nadot S., Damerval C.. (2021) Transcriptome Analysis Reveals Putative Target Genes of APETALA3-3 During Early Floral Development in Nigella damascena L.. Front. Plant Sci., (12) 660803
- Jabbour F., Pasquier PED., Chazalviel L., Guilloux ML., Conde e Silva N. , Deveaux Y., Manicacci D., Galipot P., Heiss AG., Damerval C.. (2021) Evolution of the distribution area of the Mediterranean Nigella damascena and a likely multiple molecular origin of its perianth dimorphism. Flora, (274) 151735
- Damerval C., Citerne H., Conde e Silva N. , Deveaux Y., Delannoy E., Joets J., Simonnet F., Staedler Y., Schönenberger J., Yansouni J., Le Guilloux M., Sauquet H., Nadot S.. (2019) Unraveling the Developmental and Genetic Mechanisms Underpinning Floral Architecture in Proteaceae. Front Plant Sci, (10) 18
- Denis E., Kbiri N., Mary V., Claisse G., Conde e Silva N. , Kreis M., Deveaux Y.. (2017) WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J., 3 (90) 560-572
