
Matthieu FALQUE
Recombination méiotique et évolution
- génétique végétale
- marqueurs moléculaires
- cartographie génétique
- meiose et recombinaison
- modélisation
INRAE, Ingénieur de Recherche
matthieu.falque@inrae.fr — 01 69 33 23 64
Atelier Cartographie, Expression et Polymorphisme
Biologie de l'Adaptation et Systèmes en Évolution
- Génétique Quantitative et Évolution - Le Moulon
- Université Paris-Saclay, INRAE, CNRS, AgroParisTech
- IDEEV
- 12 route 128
- 91190 Gif-sur-Yvette
Parcours
- Actuellement membre de l’équipe BASE, affilié au LABEX Saclay Plant Sciences
- HDR (habilitation à diriger les recherches) 2015, Université d’Orsay
- Responsable de l’équipe ACEP (Atelier Cartographie Expression Polymorphisme)
- Depuis 1998: Ingénieur de Recherche (INRA) at UMR GQE - Le Moulon, Gif-sur-Yvette, France
- Postdoc en génétique des populations végétales (1997-1998) CNRS, Lille France
- Postdoc en génétique des populations végétales (1995-1997) NIOO-CTO, Heteren-Wageningen, Pays-Bas
- Postdoc en génétique moléculaire végétale (1994-1995) CIRAD, Montpellier, France
- Ph.D. en biologie végétale (1994), Univ Toulouse, France
Intérêts
Mes recherches dans l’équipe BASE
2 voies de formation des crossing-overs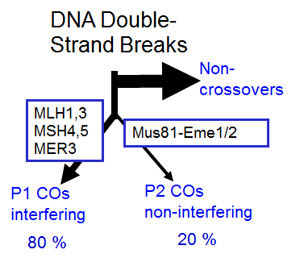
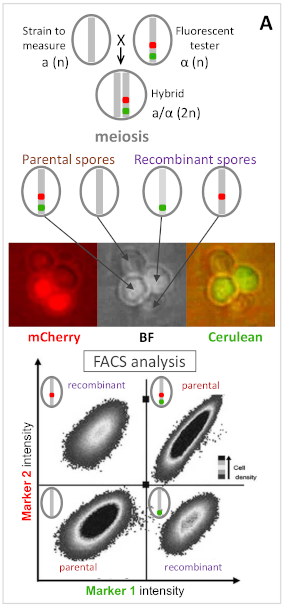
Phénotypage de la recombinaison par FACS (cas simple davec 2 marqueurs)
Plus récemment, avec Xavier Raffoux qui a effectué sa thèse avec moi, nous avons développé des approches expérimentales utilisant la levure de bière comme modèle pour étudier le déterminisme génétique de la variabilité du taux de recombinaison et de son profil le long des chromosomes.
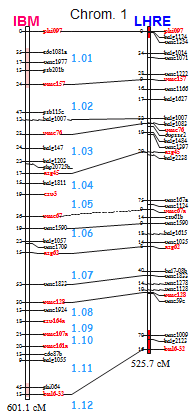
Nojs avons d’abord développé une méthode à haut-débit pour mesurer le taux de recombinaison et l’interférence grâce à des marqueurs fluorescents. Nous avons ainsi développé 8 souches “testeurs” couvrant entièrement les chromosomes VI et XI, et partiellement le chromosome I. Dans chaque testeur, trous marqueurs fluorescents ont été intégrés dans le génome à environ 30 cM l’un de l’autre, délimitant ainsi deux intervalles adjacents à partir desquels l’interférence peut être mesurée très précisément (Ref).
Nous avons ensuite appliqué cette méthode pour caractériser la diversité intra-spécifique du taux de recombinaison et de l’interférence au sein de l’espèce S. cerevisiae. Dans les différents intervalles analysés, le taux de recombinaison a montré des variations importantes entre souches, mais les profils de recombinaison le long des chromosomes varie moins. Nous avons aussi étudié le taux de recombinaison dans une région sur 10 hybrides issus de cing parents très divergents. Nos résultats indiquent globalement que le taux de recombinaison est corrélé positivement avec la similarité de séquence entre chromosomes homologues. Nous avons également estimé que les effets en cis et en trans expliquent respectivement 38% et 17% de la variance du taux de recombinaison (Ref).
Mon principal intérêt de recherche maintenant concerne les aspects évolutifs de la recombinaison méiotique. En utilisant notre méthode basée sur le FACS, nous allons faire évoluer des populations de levure sous sélection récurrente pour le taux de recombinaison, et nous utiliserons ces populations évoluées pour étudier le rôle que joue la recombinaison dans la dynamique de l’adaptation au stress.
Mes recherches dans l’équipe ACEP
L’équipe transversale ACEP est une plateforme interne de service pour la construction de cartes génétiques, les analyses de polymorphisme de l’ADN et d’expression génique. Nous fournissons un service d’analyse de marqueurs moléculaires et assurons la veille technologique sur les nouvelles technologies de marquage. Je produis aussi des cartes génétiques à haute-densité sur différentes espèces de plantes, en collaboration avec d’autres laboratoires.
Dans le cadre d’ACEP, j’ai aussi développé des outils pour :
- construire des cartes génétiques (REF) et (REF)
- détecter des CNV (Copy Number Variants) à partir de données de segrégation (REF)
- mesurer à haut-débit la cinétique de croissance de micro-organismes (REF)
- calculer automatiquement des taux de recombinaison et coefficients de coincidence à partir de données de cytométrie de flux (REF) produites avec les souches fluorescentes développées précédemment (Ref)
Projets en cours
- Dans le cadre du projet EVOLREC que je coordonne (REF) :
- Evolution expérimentale sous sélection divergent récurrente pour le taux de recombinaison méiotique
- Detection et validation fonctionnelle de QTLs de recombinaison méiotique par séquençage des populations évoluées, coll. Gianni Liti (REF)
- Evolution expérimentale sous stress salin à partir de populations avec des niveaux de recombinaison variables
- Criblage d’une banque de surexpression chez S. cerevisiae pour rechercher des modulateurs de l’interférence génétique
Activités d’enseignement
Enseignement occasionnel pour des étudiants et des sélectionneurs de sociétés privées, sur le marquage moléculaire et la cartographie génétique appliqués à l’amélioration des plantes.
Publications
- Raffoux X., Falque M. . (2025) CAYSS: Package for Automatic Cytometry Analysis of Yeast Spore Segregation. Yeast, 11-12 (41) 681-690
- Boideau F., Huteau V., Maillet L., Brunet A., Coriton O., Deniot G., Trotoux G., Taburel-Lodé M., Eber F., Gilet M., Baron C., Boutte J., Richard G., Aury JM., Belser C., Labadie K., Morice J., Falentin C., Martin O., Falque M. , Chèvre AM., Rousseau-Gueutin M.. (2024) Alternating between even and odd ploidy levels switches on and off the recombination control, even near the centromeres. The Plant Cell, 10 (36) 4472-4490
- Falque M. , Bourgais A., Dumas F., De Carvalho M., Diblasi C.. (2024) MiniRead: A simple and inexpensive do‐it‐yourself device for multiple analyses of micro‐organism growth kinetics. Yeast, yea.3932
- Bina H., Yousefzadeh H., Venon A., Remoué C., Rousselet A., Falque M. , Faramarzi S., Chen X., Samanchina J., Gill D., Kabaeva A., Giraud T., Hosseinpour B., Abdollahi H., Gabrielyan I., Nersesyan A., Cornille A.. (2022) Evidence of an additional centre of apple domestication in Iran, with contributions from the Caucasian crab apple Malus orientalis Uglitzk. to the cultivated apple gene pool. Molecular Ecology, 21 (31) 5581-5601
- Hsu YM., 2022-09, Mining genetic diversity for tomorrow's agriculture, PhD, Université Paris-Saclay
- Hsu YM., Falque M. , Martin OC.. (2022) Quantitative modelling of fine-scale variations in the Arabidopsis thaliana crossover landscape. Quant Plant Bio., (3) e3
- Vidal A., Gauthier F., Rodrigez W., Guiglielmoni N., Leroux D., Chevrolier N., Jasson S., Tourrette E., Martin OC., Falque M. . (2022) SeSAM: software for automatic construction of order-robust linkage maps. BMC Bioinformatics, 1 (23) 499
- Olvera-Vazquez SG., Alhmedi A., Miñarro M., Shykoff JA., Marchadier E., Rousselet A., Remoué C., Gardet R., Degrave A., Robert P., Chen X., Porchier J., Giraud T., Vander-Mijnsbrugee K., Raffoux X., Falque M. , Alins G., Didelot F., Beliën T., Dapena E., Lemarquand A., Cornille A.. (2021) Experimental test for local adaptation of the rosy apple aphid (Dysaphis plantaginea) to its host (Malus domestica) and to its climate in Europe. PCI Ecology, (Pre-registration version)
- Olvera-Vazquez SG., Remoué C., Venon A., Rousselet A., Grandcolas O., Azrine M., Momont L., Galan M., Benoit L., David GM., Alhmedi A., Beliën T., Alins G., Franck P., Haddioui A., Jacobsen SK., Andreev R., Simon S., Sigsgaard L., Guibert E., Tournant L., Gazel F., Mody K., Khachtib Y., Roman A., Ursu TM., Zakharov IA., Belcram H., Harry M., Roth M., Simon JC., Oram S., Ricard JM., Agnello A., Beers EH., Engelman J., Balti I., Salhi-Hannachi A., Zhang H., Tu H., Mottet C., Barrès B., Degrave A., Razmjou J., Giraud T., Falque M. , Dapena E., Miñarro M., Jardillier L., Deschamps P., Jousselin E., Cornille A.. (2021) Large-scale geography survey provides insights into the colonization history of a major aphid pest on its cultivated apple host in Europe, North America and North Africa. Peer Community Journal, (1)
- Tourrette E., Falque M. , Martin OC.. (2021) Enhancing backcross programs through increased recombination. Genet Sel Evol, 1 (53) 25
- Carrillo-Perdomo E., Vidal A., Kreplak J., Duborjal H., Leveugle M., Duarte J., Desmetz C., Deulvot C., Raffiot B., Marget P., Tayeh N., Pichon JP., Falque M. , Martin OC., Burstin J., Aubert G.. (2020) Development of new genetic resources for faba bean (Vicia faba L.) breeding through the discovery of gene-based SNP markers and the construction of a high-density consensus map. Sci Rep, 1 (10) 6790
- Falque M. , Jebreen K., Paux E., Knaak C., Mezmouk S., Martin OC.. (2020) CNVmap: A Method and Software To Detect and Map Copy Number Variants from Segregation Data. Genetics, 3 (214) 561-576
- Courret C., Gérard PR., Ogereau D., Falque M. , Moreau L., Montchamp-Moreau C.. (2019) X-chromosome meiotic drive in Drosophila simulans: a QTL approach reveals the complex polygenic determinism of Paris drive suppression. Heredity, 6 (122) 906-915
- Fustier MA., Martínez-Ainsworth NE., Aguirre-Liguori JA., Venon A., Corti H., Rousselet A., Dumas F., Dittberner H., Camarena MG., Grimanelli D., Ovaskainen O., Falque M. , Moreau L., Meaux J., Montes-Hernández S., Eguiarte LE., Vigouroux Y., Manicacci D., Tenaillon MI.. (2019) Common gardens in teosintes reveal the establishment of a syndrome of adaptation to altitude. PLOS Genetics, 12 (15) e1008512
- Kreplak J., Madoui MA., Cápal P., Novák P., Labadie K., Aubert G., Bayer PE., Gali KK., Syme RA., Main D., Klein A., Bérard A., Vrbová I., Fournier C., d’Agata L., Belser C., Berrabah W., Toegelová H., Milec Z., Vrána J., Lee HT., Kougbeadjo A., Térézol M., Huneau C., Turo CJ., Mohellibi N., Neumann P., Falque M. , Gallardo K., McGee R., Tar’an B., Bendahmane A., Aury JM., Batley J., Le Paslier MC., Ellis N., Warkentin TD., Coyne CJ., Salse J., Edwards D., Lichtenzveig J., Macas J., Doležel J., Wincker P., Burstin J.. (2019) A reference genome for pea provides insight into legume genome evolution. Nat Genet, 9 (51) 1411-1422
- Martinez Palacios P., Jacquemot MP., Tapie M., Rousselet A., Diop M., Remoué C., Falque M. , Lloyd A., Jenczewski E., Lassalle G., Chévre AM., Lelandais C., Crespi M., Brabant P., Joets J., Alix K.. (2019) Assessing the Response of Small RNA Populations to Allopolyploidy Using Resynthesized Brassica napus Allotetraploids. Mol Biol Evol, 4 (36) 709-726
- Termolino P., Falque M. , Aiese Cigliano R., Cremona G., Paparo R., Ederveen A., Martin OC., Consiglio FM., Conicella C.. (2019) Recombination suppression in heterozygotes for a pericentric inversion induces the interchromosomal effect on crossovers in Arabidopsis. Plant J, 6 (100) 1163-1175
- Tourrette E., Bernardo R., Falque M. , Martin OC.. (2019) Assessing by Modeling the Consequences of Increased Recombination in Recurrent Selection of Oryza sativa and Brassica rapa. G3, 12 (9) 4169-4181
- Tourrette E., 25 novembre 2019, Unleashing genetic diversity in breeding by increasing recombination: an in silico study, PhD, Université de Paris-Diderot
- Virlouvet L., El Hage F., Griveau Y., Jacquemot MP., Gineau E., Baldy A., Legay S., Horlow C., Combes V., Bauland C., Palafre C., Falque M. , Moreau L., Coursol S., Méchin V., Reymond M.. (2019) Water Deficit-Responsive QTLs for Cell Wall Degradability and Composition in Maize at Silage Stage. Front. Plant Sci., (10) 488
- Raffoux X., 2018-11, Diversité et déterminisme génétique de la recombinaison méiotique chez Saccharomyces cerevisiae, PhD, Université Paris Saclay (COmUE)
- Raffoux X., Bourge M., Dumas F., Martin OC., Falque M. . (2018) High-throughput measurement of recombination rates and genetic interference in Saccharomyces cerevisiae. Yeast, 6 (35) 431-442
- Raffoux X., Bourge M., Dumas F., Martin OC., Falque M. . (2018) Role of Cis, Trans, and Inbreeding Effects on Meiotic Recombination in Saccharomyces cerevisiae. Genetics, 4 (210) 1213-1226
- Giraud H., Bauland C., Falque M. , Madur D., Combes V., Jamin P., Monteil C., Laborde J., Palaffre C., Gaillard A., Blanchard P., Charcosset A., Moreau L.. (2017) Linkage Analysis and Association Mapping QTL Detection Models for Hybrids Between Multiparental Populations from Two Heterotic Groups: Application to Biomass Production in Maize (Zea mays L.). G3: Genes, Genomes, Genetics, g3.300121.2017
- Giraud H., Bauland C., Falque M. , Madur D., Combes V., Jamin P., Monteil C., Laborde J., Palaffre C., Gaillard A., Blanchard P., Charcosset A., Moreau L.. (2017) Reciprocal Genetics: Identifying QTL for General and Specific Combining Abilities in Hybrids Between Multiparental Populations from Two Maize (Zea mays L.) Heterotic Groups. Genetics, 3 (207) 1167-1180
- Pelé A., Falque M. , Trotoux G., Eber F., Nègre S., Gilet M., Huteau V., Lodé M., Jousseaume T., Dechaumet S., Morice J., Poncet C., Coriton O., Martin OC., Rousseau-Gueutin M., Chèvre AM.. (2017) Amplifying recombination genome-wide and reshaping crossover landscapes in Brassicas. PLoS Genet, 5 (13)
- Boutet G., Alves Carvalho S., Falque M. , Peterlongo P., Lhuillier E., Bouchez O., Lavaud C., Pilet-Nayel ML., Rivière N., Baranger A.. (2016) SNP discovery and genetic mapping using genotyping by sequencing of whole genome genomic DNA from a pea RIL population. BMC Genomics, (17)
- Durand E., Tenaillon MI., Raffoux X., Thépot S., Falque M. , Jamin P., Bourgais A., Ressayre A., Dillmann C.. (2015) Dearth of polymorphism associated with a sustained response to selection for flowering time in maize. BMC Evol Biol, 1 (15) 103
- Falque M. , 2015-10-15 15/10/15, Sur les traces de la recombinaison méiotique, source de biodiversité et outil génétique, HDR, Université Paris-Sud
- Sidhu GK., Fang C., Olson MA., Falque M. , Martin OC., Pawlowski WP.. (2015) Recombination patterns in maize reveal limits to crossover homeostasis. PNAS, 52 (112) 15982-15987
- Tayeh N., Aluome C., Falque M. , Jacquin F., Klein A., Chauveau A., Bérard A., Houtin H., Rond C., Kreplak J., Boucherot K., Martin C., Baranger A., Pilet‐Nayel ML., Warkentin TD., Brunel D., Marget P., Paslier MCL., Aubert G., Burstin J.. (2015) Development of two major resources for pea genomics: the GenoPea 13.2K SNP Array and a high-density, high-resolution consensus genetic map. The Plant Journal, 6 (84) 1257-1273
- Anderson LK., Lohmiller LD., Tang X., Hammond DB., Javernick L., Shearer L., Basu-Roy S., Martin OC., Falque M. . (2014) Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proceedings of the National Academy of Sciences of the United States of America, 37 (111) 13415-20
- Giraud H., Lehermeier C., Bauer E., Falque M. , Segura V., Bauland C., Camisan C., Campo L., Meyer N., Ranc N., Schipprack W., Flament P., Melchinger AE., Menz M., Moreno-Gonzalez J., Ouzunova M., Charcosset A., Schon CC., Moreau L.. (2014) Linkage disequilibrium with linkage analysis of multiline crosses reveals different multiallelic QTL for hybrid performance in the flint and dent heterotic groups of maize. Genetics, 4 (198) 1717-34
- Jahns MT., Vezon D., Chambon A., Pereira L., Falque M. , Martin OC., Chelysheva L., Grelon M.. (2014) Crossover localisation is regulated by the neddylation posttranslational regulatory pathway. PLoS biology, 8 (12) e1001930
- Roy S., 2014-03-31 31/03/14, Models of meiotic crossover formation, heterogeneity and inter-pathway cross-talks, PhD thesis, Université Paris Sud
- Suay L., Zhang DS., Eber F., Jouy H., Lode M., Huteau V., Coriton O., Szadkowski E., Leflon M., Martin OC., Falque M. , Jenczewski E., Paillard S., Chevre AM.. (2014) Crossover rate between homologous chromosomes and interference are regulated by the addition of specific unpaired chromosomes in Brassica. The New phytologist, 2 (201) 645-656
- Basu-Roy S., Gauthier F., Giraut L., Mezard C., Falque M. , Martin OC.. (2013) Hot Regions of Noninterfering Crossovers Coexist with a Nonuniformly Interfering Pathway in Arabidopsis thaliana. Genetics, 3 (195) 769-79
- Bauer E., Falque M. , Walter H., Bauland C., Camisan C., Campo L., Meyer N., Ranc N., Rincent R., Schipprack W., Altmann T., Flament P., Melchinger AE., Menz M., Moreno-Gonzalez J., Ouzunova M., Revilla P., Charcosset A., Martin OC., Schon CC.. (2013) Intraspecific variation of recombination rate in maize. Genome biology, 9 (14) R103
- Drouaud J., Khademian H., Giraut L., Zanni V., Bellalou S., Henderson IR., Falque M. , Mezard C.. (2013) Contrasted Patterns of Crossover and Non-crossover at Arabidopsis thaliana Meiotic Recombination Hotspots. PLoS genetics, 11 (9) e1003922
- Sarilar V., Martinez-Palacios P., Rousselet A., Ridel C., Falque M. , Eber F., Chevre AM., Joets J., Brabant P., Alix K.. (2013) Allopolyploidy has a moderate impact on restructuring at three contrasting transposable element insertion sites in resynthesized Brassica napus allotetraploids. The New phytologist, 2 (198) 593-604
- Ganal MW., Durstewitz G., Polley A., Bérard A., Buckler ES., Charcosset A., Clarke JD., Graner EM., Hansen M., Joets J., Le Paslier MC., McMullen MD., Montalent P., Rose M., Schön CC., Sun Q., Walter H., Martin OC., Falque M. , Lukens L.. (2011) A Large Maize (Zea mays L.) SNP Genotyping Array: Development and Germplasm Genotyping, and Genetic Mapping to Compare with the B73 Reference Genome. PLoS ONE, 12 (6) e28334
- Gauthier F., Martin O., Falque M. . (2011) CODA (crossover distribution analyzer): quantitative characterization of crossover position patterns along chromosomes. BMC Bioinformatics, 1 (12) 27
- Giraut L., Falque M. , Drouaud J., Pereira L., Martin OC., Mezard C.. (2011) Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS genetics, 11 (7) e1002354
- Rousset M., Bonnin I., Remoué C., Falque M. , Rhone B., Veyrieras JB., Madur D., Murigneux A., Balfourier F., Le Gouis J., Santoni S., Goldringer I.. (2011) Deciphering the genetics of flowering time by an association study on candidate genes in bread wheat (Triticum aestivum L.). Theor Appl Genet, 6 (123) 907-26
- Virlouvet L., Jacquemot MP., Gerentes D., Corti H., Bouton S., Gilard F., Valot B., Trouverie J., Tcherkez G., Falque M. , Damerval C., Rogowsky P., Perez P., Noctor G., Zivy M., Coursol S.. (2011) The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant physiology, 2 (157) 917-36
- Capelle V., Remoué C., Moreau L., Reyss A., Mahe A., Massonneau A., Falque M. , Charcosset A., Thevenot C., Rogowsky P., Coursol S., Prioul JL.. (2010) QTLs and candidate genes for desiccation and abscisic acid content in maize kernels. BMC plant biology, (10) 2
- Cassan L., Moreau L., Segouin S., Bellamy A., Falque M. , Limami AM.. (2010) Genetic map construction and quantitative trait loci (QTL) mapping for nitrogen use efficiency and its relationship with productivity and quality of the biennial crop Belgian endive (Cichorium intybus L.). Journal of plant physiology, 15 (167) 1253-63
- Storlazzi A., Gargano S., Ruprich-Robert G., Falque M. , David M., Kleckner N., Zickler D.. (2010) Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell, 1 (141) 94-106
- Edelist C., Raffoux X., Falque M. , Dillmann C., Sicard D., Rieseberg LH., Karrenberg S.. (2009) Differential expression of candidate salt-tolerance genes in the halophyte Helianthus paradoxus and its glycophyte progenitors H. annuus and H. petiolaris (Asteraceae). American journal of botany, 10 (96) 1830-8
- Falque M. , Anderson LK., Stack SM., Gauthier F., Martin OC.. (2009) Two types of meiotic crossovers coexist in maize. The Plant cell, 12 (21) 3915-25
- Saintenac C., Falque M. , Martin OC., Paux E., Feuillet C., Sourdille P.. (2009) Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics, 2 (181) 393-403
- Falque M. , Mercier R., Mézard C., de Vienne D., Martin OC.. (2007) Patterns of Recombination and MLH1 Foci Density Along Mouse Chromosomes: Modeling Effects of Interference and Obligate Chiasma. Genetics, 3 (176) 1453-1467