OptimaRs is a decision support tool to conduct Marker-Assisted Selection programmes.
 The main goal of OptimaRs is to help breeders in implementing their Marker-Assisted Selection (MAS) project.
With the possibility to consider a multi-allelic context, it opens new prospects to further accelerate genetic gain by assembling favorable alleles issued from diverse parents.
The main goal of OptimaRs is to help breeders in implementing their Marker-Assisted Selection (MAS) project.
With the possibility to consider a multi-allelic context, it opens new prospects to further accelerate genetic gain by assembling favorable alleles issued from diverse parents.
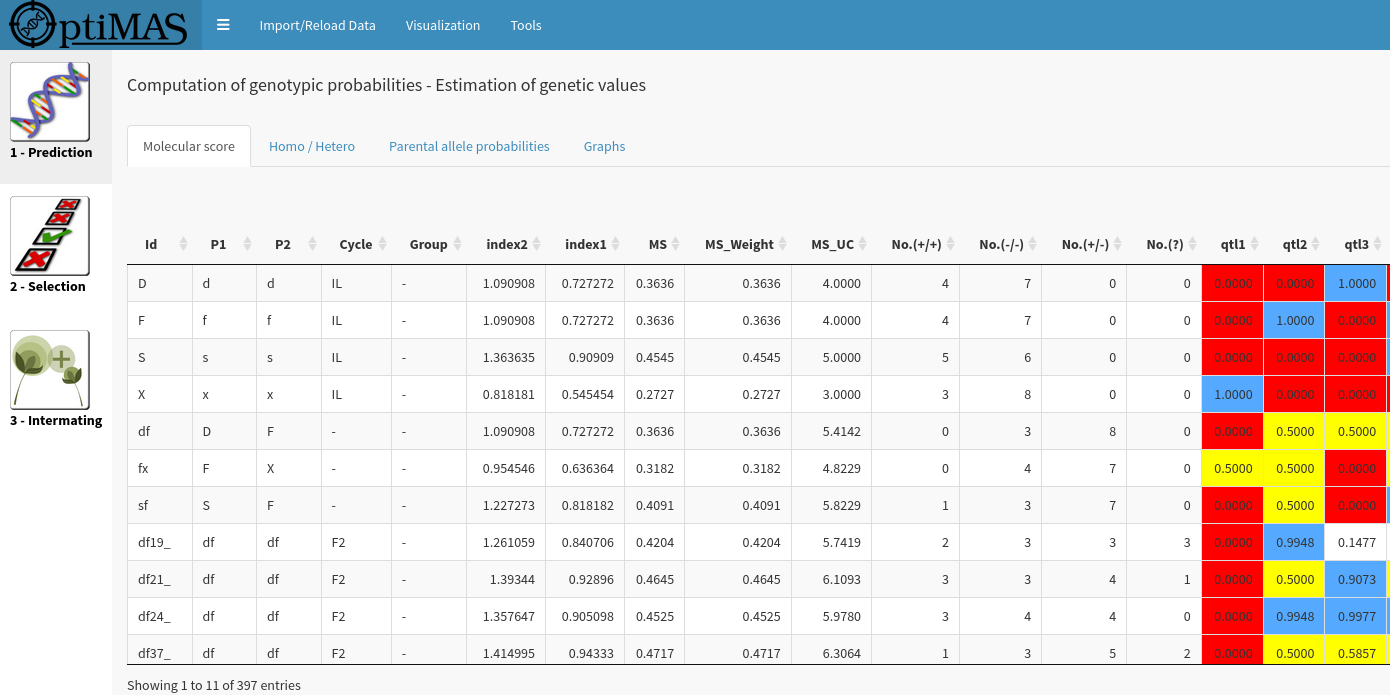
OptimaRs includes in a Graphical User Interface (GUI), three different modules corresponding to the different steps of a selection programme :
Using information given by markers located in the vicinity of the estimated QTL positions, probabilities of favorable parental QTL allele transmission are computed in different MAS schemes and mating designs (intercrossing, selfing, backcrossing, double haploids, RIL) with the possibility of considering generations without genotypic information (step 1). Then strategies are proposed to select the best plants (step 2), and efficiently intermate them based on the expected value of their progenies (step 3).