Functional validation of recombination QTLs in S.cerevisiae within the BASE team - Biology of Adaptation and Systems in Evolution.
Ingeneer (M2) position for 16 months within the ANR project EVOLREC.
May start from January 1st, 2023 on.
Scientific context and research questions
Meiotic recombination through crossing-over (CO) is a key process with major implications in genome evolution, dynamics of trait adaptation and organismal fitness. However, CO number and positionning are strongly regulated but the selective pressures that have led to these regulations remain unclear. In fact, the interplay between recombination and adaptation is complex because COs can modulate the speed of adaptation via exploiting standing genetic variation, and inversely, evolutionary processes leading to adaptation can indirectly drive changes in CO rates.
The EVOLREC project addresses the following scientific questions:
- Is recombination rate able to evolve as a response to artificial directional selection ?
- What are the CIS and TRANS genetic determinants of CO number and positioning ?
- What is the interplay between recombination and adaptation to stress ?
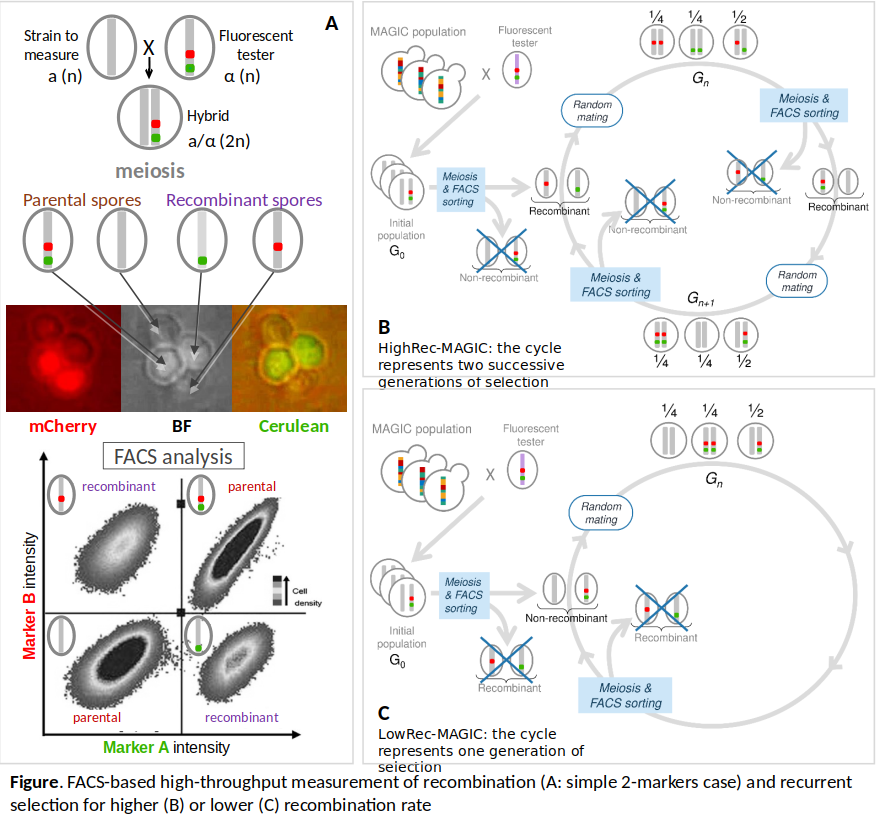
To address these questions experimentally in S. cerevisiae, we have already developed a high-throughput method based on flow cytometry using fluorescent markers, to measure recombination and sort recombinant spores by FACS [1, 2]. We have carried out 10 generations of experimental evolution under divergent recurrent selection for higher or lower recombination rate (see Figure below) in a yeast population with large genetic diversity reproducing sexually. Evolved populations have then been pool-sequenced to detect recombination QTLs (acting in cis and in trans) based on allelic frequency changes.
Experimental approaches and focus of the job
From these QTLs, candidate genes have been indentified.
Now, with the help of the person who will be recruited, we will validate the biological function of these genes on modulating recombination. To do so, we will use functional genomics approaches such as overexpression and allelic replacement. For instance, we will use homologous recombination or CRISPR-Cas9 approaches to replace the allele of the candidate gene in the low-recombining parent by the allele of the high-recombining parent, and reciprocally. Then we will measure changes in expression level and in phenotype. We will also use an overexpression library to investigate the role of candidate gene expression level on recombination. Recombination phenotypes will be measured at high-throughput by flow cytometry in several genomic regions.
All ressources - biological material, tools, and methods - necessary to the project have already been produced or obtained. Our lab is equipped with large molecular biology and microbiology facilities.
Working environment
Our lab is located in the Paris-Saclay campus, a large concentration of scientific and higher education institutions in close proximity to the Gif-sur-Yvette CNRS center, Paris-Saclay university, and at 45 minutes by train from Paris.
Key-words
molecular biology, genetic transformation, recombination, QTLs, S. cerevisiae
Required skills
Masters in molecular biology, genetics, or microbiology. Wet-lab experience is required (functional genomics or yeast experience is a plus). Spoken and written english
Contacts
Applications should be sent by e-mail to Matthieu Falque