Postdoc position (24 months): Development of a pipeline to detect polygenic selection in gene expression regulatory networks. GEvAD team hires one research engineer in population genetics (12 month contract - renewable once) as part of the NETWITS ANR
- Localisation: GEvAD team, GQE-Le Moulon, IDEEV, Gif-sur-Yvette (91)
- Duration: 12-month contract, renewable once, starting from March 2024
- Gross Salary: 2643 € - 3342€ depending on employment history

Key-words: population genetics, polygenic selection, maize adaptation, water deficit
Context and work environment
You will be welcomed in the GEvAD team L’équipe GEvAD at the UMR Quantitative genetics and Evolution (GQE) – Le Moulon (Gif-sur-Yvette, France). The team combines various approaches (experiments, computational models, population genetics, genomics, systems biology) to understand the evolutionary mechanisms behind the domestication and environmental adaptation of crops. GQE is part of Institut Diversité, Écologie, Évolution du Vivant, located on the Paris-Saclay campus. The postdoc will participate to the ANR JCJC NETWITS project, led by M. Fagny.
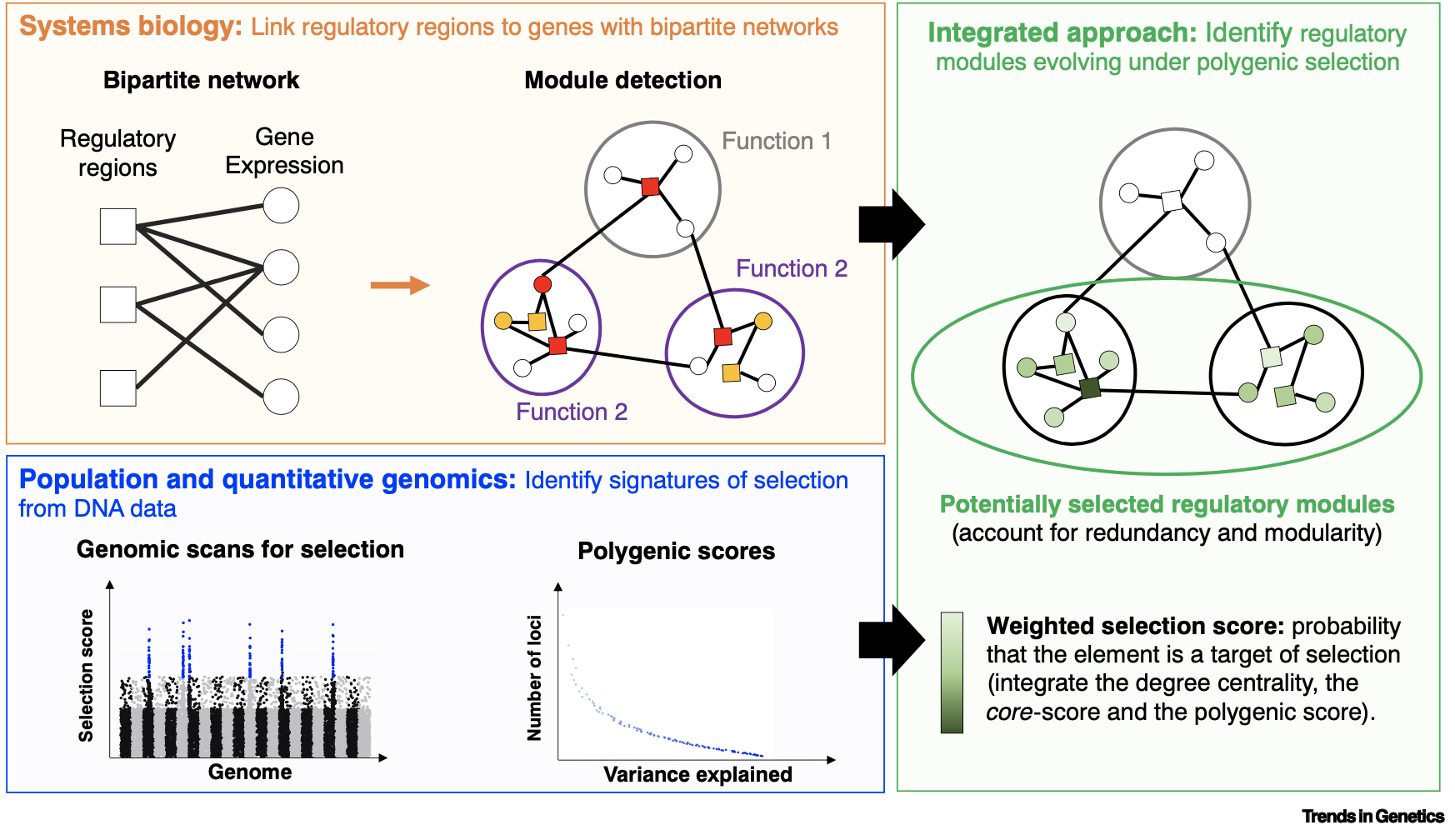
Scientific context and research questions
The NETWITS project tackles the question of the role played by gene expression regulation in maize response to water deficit. Maize tolerance to wate deficit depends strongly on its genotype. This polygenic character is determined by dozens of loci, and gene expression regulation probably plays a major role in this process. The genes involved and their regulatory regions are interacting within a complex network. The relative adaptation of maize to the temperate European climate, marked by water deficit, may have leveraged several parts of this regulatory network depending on the genetic group and population considered. However, detecting polygenic selection signatures in a regulatory network and identifying the differents paths towards adaptation is still an open question in population genetics. The postdoc, specialist in population genetics will aim to develop a pipeline to detect signatures of polygenic selection in the context of epistasis and pleiotropy, in order to study the link between gene regulatory network topology and natural selection. The postdoc will use maize adaptation to water deficit as a model. The postdoc will collaborate closely with another Postdoctoral fellow who will be hired as part of the NETWITS project.
Missions
- Simulate various polygenic selection scenarios targeting gene expression regulation;
- Develop a pipeline to detect polygenic selection signatures;
- Test the statistical power of this pipeline and its sensitivity to various confounding factors;
- Apply this pipeline to genomic and transcriptomic data from maize grown with and without water deficit;
- Prepare data for publication in publicly available repository (e.g.: dataverse INRAE);
- Contribute to writing articles.
Required skills and knowledges
- PhD ;
- Recommended training: Theoretical and applied population genetics.
- Required skills and knowledges: - Forward-in-time population evolution models, summary statistics computation. - Bioinformatics: programming skills in python, shell, R, SLiM, FAIR data management, knows how to use computational clusters; - Scientific articles writing; - Spoken & written English: B1 to B2 level (Common European Framework of Reference for Languages);
- Knowledges in systems biology/gene regulatory networks will be appreciated but are not necessary.
To apply
Send a CV, a cover letter, and the contact information for 2 referees to maud.fagny@inrae.fr.