PAPPSO - Plateforme d'Analyse Protéomique de Paris Sud-Ouest
La plateforme PAPPSO est une installation collective dédiée à l’analyse protéomique. Elle est accessible aux unités de recherche académique et aux utilisateurs privés dans le cadre de prestations et de collaborations.
Missions
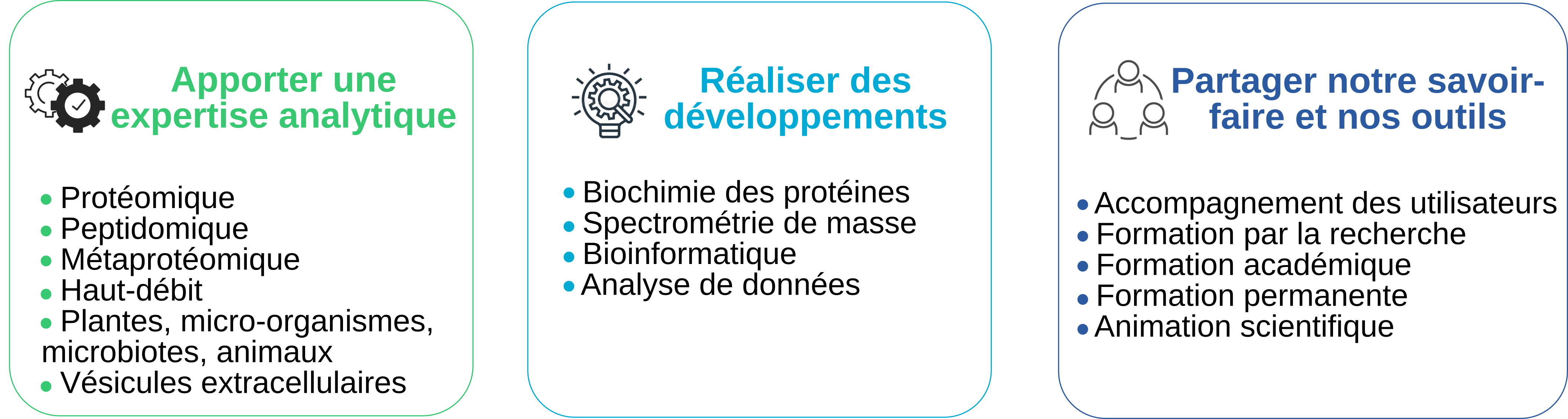
L’objectif de PAPPSO est de mettre à disposition de ses utilisateurs un équipement, une compétence scientifique et un savoir-faire en protéomique par spectrométrie de masse qui soient les mieux adaptés à leurs besoins. Les missions de PAPPSO se déclinent en trois volets :
Spécificités
PAPPSO est spécialisée dans l’analyse protéomique par spectrométrie de masse. Cette technologie permet d’identifier, de quantifier les protéines et d’analyser leurs modifications post-traduictionnelles. Dans le cadre des projets de recherche dans lesquels elle est impliquée, PAPPSO est régulièrement amenée à réaliser des travaux de recherche et de développement grâce auxquels elle présente aujourd’hui les spécificités suivantes :
- Analyse de gros dispositifs expérimentaux incluant jusqu’à plusieurs centaines échantillons pouvant être très complexes ;
- Caractérisation de modifications post-traductionnelles (principalement phosphorylations mais aussi glycosylations, acétylations, méthylations) à l’aide de méthodologies appropriées ;
- Expertise dans l’analyse protéomique des plantes, des animaux et des micro-organismes (incluant les microbiomes humain et du sol) en cohérence avec les recherches menées dans les deux unités d’adossement ;
- Analyse de mélanges de peptides natifs potentiellement porteurs de modifications post-traductionnelles originales ;
- Développement de logiciels de bio-informatique en constante évolution et couvrant tous les aspects de l’analyse protéomique depuis l’exploitation des données brutes de spectromètrie de masse jusqu’aux analyses statistiques. Ces logiciels sont adaptés aux très gros volumes de données et permettent de traiter simultanément les données issues de centaines d’échantillons très complexes. Ils sont publiés sous licence libre GPL et sont utilisés globalement, y compris à l’étranger.
Organisation
PAPPSO est organisée en deux plateaux techniques complémentaires, fonctionnant grâce à des personnels compétents et formés en biochimie des protéines, spectrométrie de masse, bio-informatique et statistiques :
- PAPPSO Moulon : adossé à l’UMR GQE-Le Moulon, ce site est spécialisé dans la préparation et l’analyse d’échantillons végétaux;
- PAPPSO Jouy : adossé à l’UMR Micalis (Jouy-en-Josas), ce site est spécialisé dans l’analyse d’échantillons animaux et microbiens (notamment le microbiote intestinal humain).

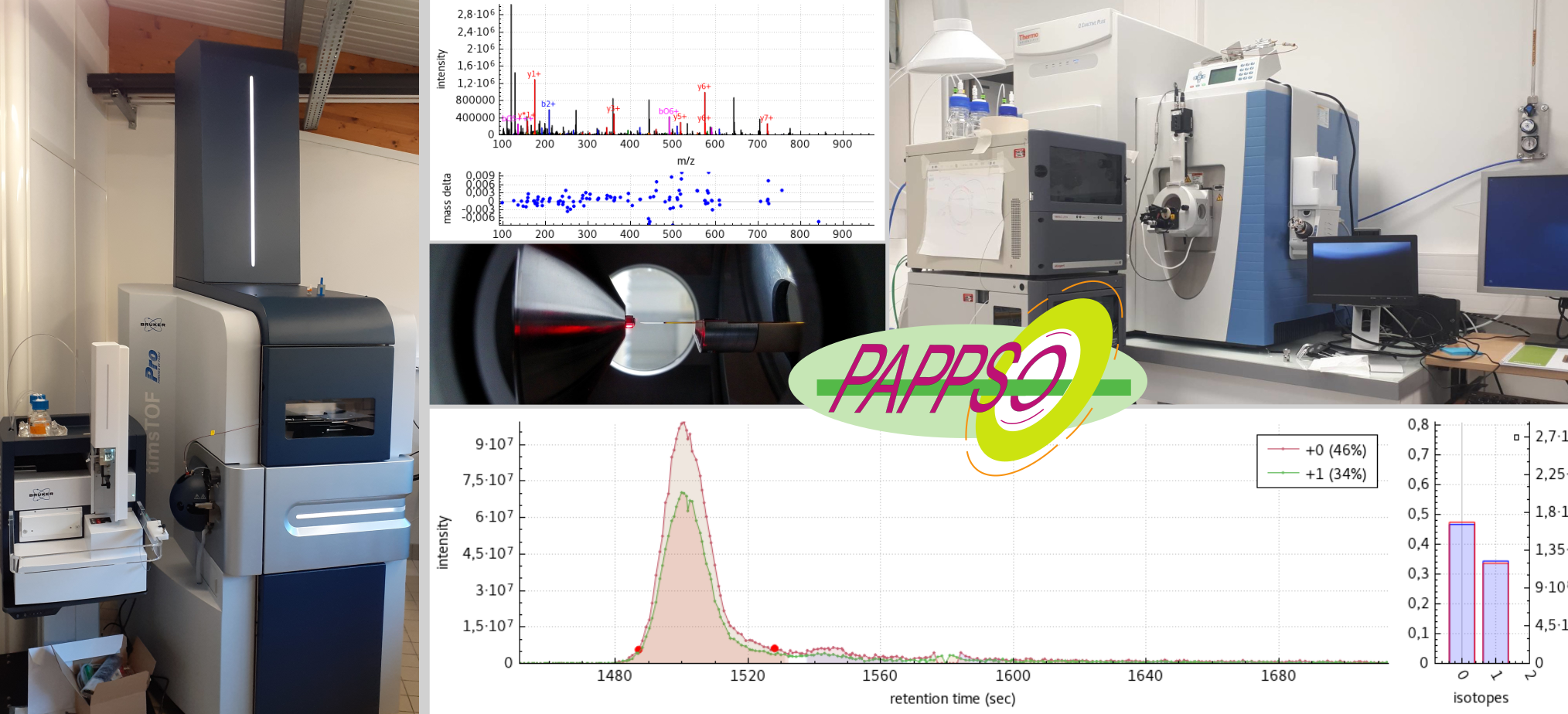
Equipement
PAPPSO est équipée de trois spectromètres de masse à haute résolution couplés à des appareils de chromatographie liquide à haute performance, dont deux sur le site PAPPSO Jouy et un sur le site PAPPSO Moulon. La plateforme est également dotée d’une infrastructure informatique qui lui permet de gérer de très gros volumes de données.

Labels et certification
PAPPSO est labelisée par IBiSA et par INRAE en tant qu’installation scientifique collective (ISC). Elle est également certifiée ISO 9001:2015 depuis 2017.

Membres
- Thierry Balliau Assistant Ingénieur (INRAE)
- Willy Vincent Bienvenut Chargé de recherche (CNRS), 50% BASE, 50% PAPPSO
- Mélisande Blein-Nicolas Ingénieure de Recherche (INRAE), 50% BASE, 50% PAPPSO
- Marlène Davanture (CNRS)
- Laurencia Fera (INRAE)
- Olivier Langella Ingénieur de Recherche (CNRS), 10% BiOSS, 90% PAPPSO
- Pierre Leriche Technicien (INRAE)
- Filippo Rusconi (CNRS)
Anciens membres
Guillaume Sarrailhé Ingénieur d'étude (2023-2024), Michel Zivy Directeur de Recherche (1985-2022), Luciana De Oliveira Postdoc (2020-2021), Thomas Renne Apprenti en Bioinformatique (2019-2021), Benoît Valot Ingénieur de Recherche (2006-2014)Publications
- Balliau T. , Frambourg A., Langella O. , Martin ML., Zivy M., Blein-Nicolas M. . (2025) MCQR : Enhancing the Processing and Analysis of Quantitative Proteomics Data by Incorporating Chromatography and Mass Spectrometry Information. J. Proteome Res., acs.jproteome.4c01119
- Andrieu, Marie-Hélène ., Atienza, Isabelle ., Marmagne, Anne ., Just, Daniel ., Bienvenut, Willy Vincent ., Gibon, Yves ., Colombié, Sophie .. (2024) Protocole de marquage à l’azote 15N de fruits de tomate issus de plantes cultivées en condition standard. NOV'AE, 5 (2022) 1-11
- Balliau T. , Ashenafi M., Blein-Nicolas M. , Turc O., Zivy M., Marchadier E.. (2024) A Moderate Water Deficit Induces Profound Changes in the Proteome of Developing Maize Ovaries. Biomolecules, 10 (14) 1239
- Blein-Nicolas M. , Devijver E., Gallopin M., Perthame E.. (2024) Nonlinear network-based quantitative trait prediction from biological data. Journal of the Royal Statistical Society Series C: Applied Statistics, qlae012
- Langella O. , Renne T., Balliau T. , Davanture M. , Brehmer S., Zivy M., Blein-Nicolas M. , Rusconi F. . (2024) Full Native timsTOF PASEF-Enabled Quantitative Proteomics with the i2MassChroQ Software Package. J. Proteome Res., 8 (23) 3353-3366
- Tarkowski Łukasz P., Clochard T., Blein-Nicolas M. , Zivy M., Baillau T., Abadie C., Morère-Le Paven MC., Limami AM., Tcherkez G., Montrichard F.. (2024) The nitrate transporter-sensor MtNPF6.8 regulates the branched chain amino acid/pantothenate metabolic pathway in barrel medic (Medicago truncatula Gaertn.) root tip. Plant Physiology and Biochemistry, (206) 108213
- Urrutia M., Blein-Nicolas M. , Fernandez O., Bernillon S., Maucourt M., Deborde C., Balliau T. , Rabier D., Bénard C., Prigent S., Quilleré I., Jacob D., Gibon Y., Zivy M., Giauffret C., Hirel B., Moing A.. (2024) Identification of metabolic and protein markers representative of the impact of mild nitrogen deficit on agronomic performance of maize hybrids. Metabolomics, 6 (20) 128
- Chasseriaud L., Albertin W., Blein-Nicolas M. , Balliau T. , Zivy M., Coulon J., Bely M.. (2023) Physical Contact between Torulaspora delbrueckii and Saccharomyces cerevisiae Alters Cell Growth and Molecular Interactions in Grape Must. Beverages, 3 (9) 81
- Duruflé H., Balliau T. , Blanchet N., Chaubet A., Duhnen A., Pouilly N., Blein-Nicolas M. , Mangin B., Maury P., Langlade NB., Zivy M.. (2023) Sunflower Hybrids and Inbred Lines Adopt Different Physiological Strategies and Proteome Responses to Cope with Water Deficit. Biomolecules, 7 (13) 1110
- Jamet E., Esquerré-Tugayé MT., Gallardo-Guerrero K., Rolland N., Zivy M., Blein-Nicolas M. , Vincent D., Gontero B., Rajjou L.. (2023) Obituary: Dominique Job (1947-2022). Front. Plant Sci., (14) 1188766
- Prunier G., Cherkaoui M., Lysiak A., Langella O. , Blein-Nicolas M. , Lollier V., Benoist E., Jean G., Fertin G., Rogniaux H., Tessier D.. (2023) Fast alignment of mass spectra in large proteomics datasets, capturing dissimilarities arising from multiple complex modifications of peptides. BMC Bioinformatics, 1 (24) 421
- Atze H., Liang Y., Hugonnet JE., Gutierrez A., Rusconi F. , Arthur M.. (2022) Heavy isotope labeling and mass spectrometry reveal unexpected remodeling of bacterial cell wall expansion in response to drugs. eLife, (11) e72863
- Baudouin E., Puyaubert J., Meimoun P., Blein-Nicolas M. , Davanture M. , Zivy M., Bailly C.. (2022) Dynamics of Protein Phosphorylation during Arabidopsis Seed Germination. International Journal of Molecular Sciences, 13 (23) 7059
- Guillou MC., Balliau T. , Vergne E., Canut H., Chourré J., Herrera-León C., Ramos-Martín F., Ahmadi-Afzadi M., D'Amelio N., Ruelland E., Zivy M., Renou JP., Jamet E., Aubourg S.. (2022) The PROSCOOP10 Gene Encodes Two Extracellular Hydroxylated Peptides and Impacts Flowering Time in Arabidopsis. Plants (Basel), 24 (11) 3554
- Henry C., Bassignani A., Berland M., Langella O. , Sokol H., Juste C.. (2022) Modern Metaproteomics: A Unique Tool to Characterize the Active Microbiome in Health and Diseases, and Pave the Road towards New Biomarkers—Example of Crohn’s Disease and Ulcerative Colitis Flare-Ups. Cells, 8 (11) 1340
- Jean N., Perié L., Dumont E., Bertheau L., Balliau T. , Caruana AMN., Amzil Z., Laabir M., Masseret E.. (2022) Metal stresses modify soluble proteomes and toxin profiles in two Mediterranean strains of the distributed dinoflagellate Alexandrium pacificum. Sci Total Environ, (818) 151680
- Kalemba EM., Valot B., Job D., Bailly C., Meimoun P.. (2022) Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsis thaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis. Plants, 4 (11) 569
- Millán-Oropeza A., Blein-Nicolas M. , Monnet V., Zivy M., Henry C.. (2022) Comparison of Different Label-Free Techniques for the Semi-Absolute Quantification of Protein Abundance. Proteomes, 1 (10) 2
- Plancade S., Berland M., Blein-Nicolas M. , Langella O. , Bassignani A., Juste C.. (2022) A combined test for feature selection on sparse metaproteomics data—an alternative to missing value imputation. PeerJ, (10) e13525
- Sano N., Lounifi I., Cueff G., Collet B., Clément G., Balzergue S., Huguet S., Valot B., Galland M., Rajjou L.. (2022) Multi-Omics Approaches Unravel Specific Features of Embryo and Endosperm in Rice Seed Germination. Front. Plant Sci., (13) 867263
- Wang X., Davanture M. , Zivy M., Bailly C., Nambara E., Corbineau F.. (2022) Label-Free Quantitative Proteomics Reveal the Involvement of PRT6 in Arabidopsis thaliana Seed Responsiveness to Ethylene. International Journal of Molecular Sciences, 16 (23) 9352
- Yip Delormel T., Avila-Ospina L., Davanture M. , Zivy M., Lang J., Valentin N., Rayapuram N., Hirt H., Colcombet J., Boudsocq M.. (2022) In vivo identification of putative CPK5 substrates in Arabidopsis thaliana. Plant Science, (314) 111121
- Zang L., Tarkowski LP., Morère-Le Paven MC., Zivy M., Balliau T. , Clochard T., Bahut M., Balzergue S., Pelletier S., Landès C., Limami AM., Montrichard F.. (2022) The Nitrate Transporter MtNPF6.8 Is a Master Sensor of Nitrate Signal in the Primary Root Tip of Medicago truncatula. Front Plant Sci, (13) 832246
- Balliau T. , Duruflé H., Blanchet N., Blein-Nicolas M. , Langlade NB., Zivy M.. (2021) Proteomic data from leaves of twenty-four sunflower genotypes under water deficit. OCL, (28) 12
- Bassignani A., Plancade S., Berland M., Blein-Nicolas M. , Guillot A., Chevret D., Moritz C., Huet S., Rizkalla S., Clément K., Doré J., Langella O. , Juste C.. (2021) Benefits of Iterative Searches of Large Databases to Interpret Large Human Gut Metaproteomic Data Sets. J. Proteome Res.,
- Ben Salah E., Barrera C., Sakly W., Mosbahi S., Balliau T. , Franche N., Gottstein B., Ben Youssef S., Mekki M., Babba H., Millon L.. (2021) Novel biomarkers for the early prediction of pediatric cystic echinococcosis post-surgical outcomes. J Infect, S0163-4453(21)00494-1
- Cui J., Davanture M. , Lamade E., Zivy M., Tcherkez G.. (2021) Plant low-K responses are partly due to Ca prevalence and the low-K biomarker putrescine does not protect from Ca side effects but acts as a metabolic regulator. Plant Cell Environ, 5 (44) 1565-1579
- Douché T., Valot B., Balliau T. , San Clemente H., Zivy M., Jamet E.. (2021) Cell wall proteomic datasets of stems and leaves of Brachypodium distachyon. Data in Brief, (35) 106818
- Duminil P., Davanture M. , Oury C., Boex‐Fontvieille E., Tcherkez G., Zivy M., Hodges M., Glab N.. (2021) Arabidopsis thaliana 2,3‐bisphosphoglycerate‐independent phosphoglycerate mutase 2 activity requires serine 82 phosphorylation. Plant J, 5 (107) 1478-1489
- Henriet C., Balliau T. , Aimé D., Le Signor C., Kreplak J., Zivy M., Gallardo K., Vernoud V.. (2021) Proteomics of developing pea seeds reveals a complex antioxidant network underlying the response to sulfur deficiency and water stress. Journal of Experimental Botany, eraa571
- Kolkas H., Balliau T. , Chourré J., Zivy M., Canut H., Jamet E.. (2021) The Cell Wall Proteome of Marchantia polymorpha Reveals Specificities Compared to Those of Flowering Plants. Front Plant Sci, (12) 765846
- Langella O. , Rusconi F. . (2021) mineXpert2 : Full-Depth Visualization and Exploration of MS n Mass Spectrometry Data. J. Am. Soc. Mass Spectrom., 4 (32) 1138-1141
- Nicoud Q., Lamouche F., Chaumeret A., Balliau T. , Le Bars R., Bourge M., Pierre F., Guérard F., Sallet E., Tuffigo S., Pierre O., Dessaux Y., Gilard F., Gakière B., Nagy I., Kereszt A., Zivy M., Mergaert P., Gourion B., Alunni B.. (2021) Bradyrhizobium diazoefficiens USDA110 Nodulation of Aeschynomene afraspera Is Associated with Atypical Terminal Bacteroid Differentiation and Suboptimal Symbiotic Efficiency. mSystems, 3 (6) e01237-20
- Recorbet G., Calabrese S., Balliau T. , Zivy M., Wipf D., Boller T., Courty PE.. (2021) Proteome adaptations under contrasting soil phosphate regimes of Rhizophagus irregularis engaged in a common mycorrhizal network. Fungal Genetics and Biology, (147) 103517
- Rusconi F. . (2021) Free Open Source Software for Protein and Peptide Mass Spectrometry- based Science. CPPS, 2 (22) 134-147
- Urrutia M., Blein-Nicolas M. , Prigent S., Bernillon S., Deborde C., Balliau T. , Maucourt M., Jacob D., Ballias P., Bénard C., Sellier H., Gibon Y., Giauffret C., Zivy M., Moing A.. (2021) Maize metabolome and proteome responses to controlled cold stress partly mimic early-sowing effects in the field and differ from those of Arabidopsis. Plant Cell Environ,
- Van Den Bossche T., Kunath BJ., Schallert K., Schäpe SS., Abraham PE., Armengaud J., Arntzen M., Bassignani A., Benndorf D., Fuchs S., Giannone RJ., Griffin TJ., Hagen LH., Halder R., Henry C., Hettich RL., Heyer R., Jagtap P., Jehmlich N., Jensen M., Juste C., Kleiner M., Langella O. , Lehmann T., Leith E., May P., Mesuere B., Miotello G., Peters SL., Pible O., Queiros PT., Reichl U., Renard BY., Schiebenhoefer H., Sczyrba A., Tanca A., Trappe K., Trezzi JP., Uzzau S., Verschaffelt P., Von Bergen M., Wilmes P., Wolf M., Martens L., Muth T.. (2021) Critical Assessment of MetaProteome Investigation (CAMPI): a multi-laboratory comparison of established workflows. Nat Commun, 1 (12) 7305
- Belouah I., Bénard C., Denton A., Blein-Nicolas M. , Balliau T. , Teyssier E., Gallusci P., Bouchez O., Usadel B., Zivy M., Gibon Y., Colombié S.. (2020) Transcriptomic and proteomic data in developing tomato fruit. Data in Brief, (28) 105015
- Blein-Nicolas M. , Negro SS., Balliau T. , Welcker C., Cabrera-Bosquet L., Nicolas SD., Charcosset A., Zivy M.. (2020) A systems genetics approach reveals environment-dependent associations between SNPs, protein coexpression, and drought-related traits in maize. Genome Res., 11 (30) 1593-1604
- Chetouhi C., Masseret E., Satta CT., Balliau T. , Laabir M., Jean N.. (2020) Intraspecific variability in membrane proteome, cell growth, and morphometry of the invasive marine neurotoxic dinoflagellate Alexandrium pacificum grown in metal-contaminated conditions. Science of The Total Environment, (715) 136834
- Kilani J., Davanture M. , Simon A., Zivy M., Fillinger S.. (2020) Comparative quantitative proteomics of osmotic signal transduction mutants in Botrytis cinerea explain mutant phenotypes and highlight interaction with cAMP and Ca2+ signalling pathways. Journal of Proteomics, (212) 103580
- Luo J., Havé M., Clément G., Tellier F., Balliau T. , Launay-Avon A., Guérard F., Zivy M., Masclaux-Daubresse C.. (2020) Integrating multiple omics to dissect the common and specific molecular changes occurring in Arabidopsis thaliana (L.) under nitrate and sulfate chronic limitations. Journal of Experimental Botany, eraa337
- Saux M., Ponnaiah M., Langlade N., Zanchetta C., Balliau T. , El‐Maarouf‐Bouteau H., Bailly C.. (2020) A multiscale approach reveals regulatory players of water stress responses in seeds during germination. Plant Cell Environ, 5 (43) 1300-1313
- Tcherkez G., Carroll A., Abadie C., Mainguet S., Davanture M. , Zivy M.. (2020) Protein synthesis increases with photosynthesis via the stimulation of translation initiation. Plant Science, (291) 110352
- Bancel E., Bonnot T., Davanture M. , Alvarez D., Zivy M., Martre P., Déjean S., Ravel C.. (2019) Proteomic Data Integration Highlights Central Actors Involved in Einkorn (Triticum monococcum ssp. monococcum) Grain Filling in Relation to Grain Storage Protein Composition. Front. Plant Sci., (10) 832
- Belouah I., Nazaret C., Pétriacq P., Prigent S., Bénard C., Mengin V., Blein-Nicolas M. , Denton AK., Balliau T. , Augé S., Bouchez O., Mazat JP., Stitt M., Usadel B., Zivy M., Beauvoit B., Gibon Y., Colombié S.. (2019) Modeling Protein Destiny in Developing Fruit. Plant Physiology, 3 (180) 1709-1724
- Belouah I., Blein-Nicolas M. , Balliau T. , Gibon Y., Zivy M., Colombié S.. (2019) Peptide filtering differently affects the performances of XIC-based quantification methods. J Proteomics, (193) 131-141
- Bienvenut W. V. , 2 décembre 2019, Des débuts de la protéomique à l'acétylation N-terminale des protéines chez les plantes, HDR, Université Paris Saclay
- Chauffour F., Bailly M., Perreau F., Cueff G., Suzuki H., Collet B., Frey A., Clément G., Soubigou-Taconnat L., Balliau T. , Krieger-Liszkay A., Rajjou L., Marion-Poll A.. (2019) Multi-omics Analysis Reveals Sequential Roles for ABA during Seed Maturation. Plant Physiol., 2 (180) 1198-1218
- Cui J., Davanture M. , Zivy M., Lamade E., Tcherkez G.. (2019) Metabolic responses to potassium availability and waterlogging reshape respiration and carbon use efficiency in oil palm. New Phytologist, 1 (223) 310-322
- Duruflé H., Ranocha P., Balliau T. , Dunand C., Jamet E.. (2019) Transcriptomic and cell wall proteomic datasets of rosettes and floral stems from five Arabidopsis thaliana ecotypes grown at optimal or sub-optimal temperature. Data in Brief, (27) 104581
- Havé M., Luo J., Tellier F., Balliau T. , Cueff G., Chardon F., Zivy M., Rajjou L., Cacas JL., Masclaux‐Daubresse C.. (2019) Proteomic and lipidomic analyses of the Arabidopsis atg5 autophagy mutant reveal major changes in ER and peroxisome metabolisms and in lipid composition. New Phytol, 3 (223) 1461-1477
- Henry C., Haller L., Blein-Nicolas M. , Zivy M., Canette A., Verbrugghe M., Mézange C., Boulay M., Gardan R., Samson S., Martin V., André-Leroux G., Monnet V.. (2019) Identification of Hanks-Type Kinase PknB-Specific Targets in the Streptococcus thermophilus Phosphoproteome. Front. Microbiol., (10) 1329
- Rusconi F. . (2019) mineXpert: Biological Mass Spectrometry Data Visualization and Mining with Full JavaScript Ability. J. Proteome Res., 5 (18) 2254-2259
- Aloui A., Recorbet G., Lemaître-Guillier C., Mounier A., Balliau T. , Zivy M., Wipf D., Dumas-Gaudot E.. (2018) The plasma membrane proteome of Medicago truncatula roots as modified by arbuscular mycorrhizal symbiosis. Mycorrhiza, 1 (28) 1-16
- Balliau T. , Blein-Nicolas M. , Zivy M.. (2018) Evaluation of Optimized Tube-Gel Methods of Sample Preparation for Large-Scale Plant Proteomics. Proteomes, 1 (6) 6
- Berrabah F., Balliau T. , Ait-Salem EH., George J., Zivy M., Ratet P., Gourion B.. (2018) Control of the ethylene signaling pathway prevents plant defenses during intracellular accommodation of the rhizobia. The New phytologist, 1 (219) 310-323
- Bichang'a G., Da Lage JL., Capdevielle-Dulac C., Zivy M., Balliau T. , Sambai K., Le Ru B., Kaiser L., Juma G., Maina ENM., Calatayud PA.. (2018) alpha-Amylase Mediates Host Acceptance in the Braconid Parasitoid Cotesia flavipes. J Chem Ecol, 11 (44) 1030-1039
- Havé M., Balliau T. , Cottyn-Boitte B., Dérond E., Cueff G., Soulay F., Lornac A., Reichman P., Dissmeyer N., Avice JC., Gallois P., Rajjou L., Zivy M., Masclaux-Daubresse C.. (2018) Increases in activity of proteasome and papain-like cysteine protease in Arabidopsis autophagy mutants: back-up compensatory effect or cell-death promoting effect?. J Exp Bot, 6 (69) 1369-1385
- Bonnot T., Bancel E., Alvarez D., Davanture M. , Boudet J., Pailloux M., Zivy M., Ravel C., Martre P.. (2017) Grain subproteome responses to nitrogen and sulfur supply in diploid wheat Triticum monococcum ssp. monococcum. The Plant Journal, 5 (91) 894-910
- Duruflé H., Hervé V., Ranocha P., Balliau T. , Zivy M., Chourré J., San Clemente H., Burlat V., Albenne C., Déjean S., Jamet E., Dunand C.. (2017) Cell wall modifications of two Arabidopsis thaliana ecotypes, Col and Sha, in response to sub-optimal growth conditions: An integrative study. Plant Science, (263) 183-193
- Duruflé H., Clemente HS., Balliau T. , Zivy M., Dunand C., Jamet E.. (2017) Cell wall proteome analysis of Arabidopsis thaliana mature stems. PROTEOMICS, 8 (17) 1600449
- Duruflé H., Hervé V., Balliau T. , Zivy M., Dunand C., Jamet E.. (2017) Proline Hydroxylation in Cell Wall Proteins: Is It Yet Possible to Define Rules?. Front. Plant Sci., (8)
- Galland M., He D., Lounifi I., Arc E., Clément G., Balzergue S., Huguet S., Cueff G., Godin B., Collet B., Granier F., Morin H., Tran J., Valot B., Rajjou L.. (2017) An Integrated “Multi-Omics” Comparison of Embryo and Endosperm Tissue-Specific Features and Their Impact on Rice Seed Quality. Front Plant Sci, (8)
- Jean N., Dumont E., Herzi F., Balliau T. , Laabir M., Masseret E., Mounier S.. (2017) Modifications of the soluble proteome of a mediterranean strain of the invasive neurotoxic dinoflagellate Alexandrium catenella under metal stress conditions. Aquatic Toxicology, (188) 80-91
- Langella O. , Valot B., Balliau T. , Blein-Nicolas M. , Bonhomme L., Zivy M.. (2017) X!TandemPipeline: A Tool to Manage Sequence Redundancy for Protein Inference and Phosphosite Identification. J. Proteome Res., 2 (16) 494-503
- Millan-Oropeza A., Henry C., Blein-Nicolas M. , Aubert-Frambourg A., Moussa F., Bleton J., Virolle MJ.. (2017) Quantitative Proteomics Analysis Confirmed Oxidative Metabolism Predominates in Streptomyces coelicolor versus Glycolytic Metabolism in Streptomyces lividans. J. Proteome Res., 7 (16) 2597-2613
- Negroni L., Zivy M., Lemaire C.. (2017) Mass Spectrometry of Mitochondrial Membrane Protein Complexes. Springer Link, 233-246
- Regente M., Pinedo M., San Clemente H., Balliau T. , Jamet E., de la Canal L.. (2017) Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J Exp Bot, 20 (68) 5485-5495
- Renvoisé M., Bonhomme L., Davanture M. , Zivy M., Lemaire C.. (2017) Phosphoproteomic Analysis of Isolated Mitochondria in Yeast. Springer Link, 283-299
- Rouzet A., Reboux G., Dalphin JC., Gondouin A., De Vuyst P., Balliau T. , Millon L., Valot B., Roussel S.. (2017) An immunoproteomic approach revealed antigenic proteins enhancing serodiagnosis performance of bird fancier's lung. Journal of Immunological Methods, (450) 58-65
- Villegente M., Marmey P., Job C., Galland M., Cueff G., Godin B., Rajjou L., Balliau T. , Zivy M., Fogliani B., Sarramegna-Burtet V., Job D.. (2017) A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda. Proteomes, 3 (5)
- Blein-Nicolas M. , Zivy M.. (2016) Thousand and one ways to quantify and compare protein abundances in label-free bottom-up proteomics. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 8 (1864) 883-895
- Dobrenel T., Mancera-Martínez E., Forzani C., Azzopardi M., Davanture M. , Moreau M., Schepetilnikov M., Chicher J., Langella O. , Zivy M., Robaglia C., Ryabova LA., Hanson J., Meyer C.. (2016) The Arabidopsis TOR Kinase Specifically Regulates the Expression of Nuclear Genes Coding for Plastidic Ribosomal Proteins and the Phosphorylation of the Cytosolic Ribosomal Protein S6. Front Plant Sci, (7)
- Herve V., Durufle H., San Clemente H., Albenne C., Balliau T. , Zivy M., Dunand C., Jamet E.. (2016) An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics, 24 (16) 3183-3187
- Sabarly V., Aubron C., Glodt J., Balliau T. , Langella O. , Chevret D., Rigal O., Bourgais A., Picard B., de Vienne D., Denamur E., Bouvet O., Dillmann C.. (2016) Interactions between genotype and environment drive the metabolic phenotype within Escherichia coli isolates. Environmental microbiology, 1 (18) 100-17
- Blein-Nicolas M. , Albertin W., da Silva T., Valot B., Balliau T. , Masneuf-Pomarède I., Bely M., Marullo P., Sicard D., Dillmann C., de Vienne D., Zivy M.. (2015) A Systems Approach to Elucidate Heterosis of Protein Abundances in Yeast. Mol. Cell Proteomics, 8 (14) 2056-2071
- Hummel M., Dobrenel T., Cordewener JJ., Davanture M. , Meyer C., Smeekens SJ., Bailey-Serres J., America TA., Hanson J.. (2015) Proteomic LC-MS analysis of Arabidopsis cytosolic ribosomes: Identification of ribosomal protein paralogs and re-annotation of the ribosomal protein genes. Journal of proteomics, (128) 436-49
- Albertin W., Marullo P., Bely M., Aigle M., Bourgais A., Langella O. , Balliau T. , Chevret D., Valot B., da Silva T., Dillmann C., de Vienne D., Sicard D.. (2013) Linking post-translational modifications and variation of phenotypic traits. Molecular & cellular proteomics : MCP, 3 (12) 720-35
- Blein-Nicolas M. , Albertin W., Valot B., Marullo P., Sicard D., Giraud C., Huet S., Bourgais A., Dillmann C., de Vienne D., Zivy M.. (2013) Yeast proteome variations reveal different adaptive responses to grape must fermentation. Molecular biology and evolution, 6 (30) 1368-83
- Langella O. , Valot B., Jacob D., Balliau T. , Flores R., Hoogland C., Joets J., Zivy M.. (2013) Management and dissemination of MS proteomic data with PROTICdb: Example of a quantitative comparison between methods of protein extraction. Proteomics, 9 (13) 1457-66
- Blein-Nicolas M. , Xu H., de Vienne D., Giraud C., Huet S., Zivy M.. (2012) Including shared peptides for estimating protein abundances: a significant improvement for quantitative proteomics. Proteomics, 18 (12) 2797-801
- Valot B., Langella O. , Nano E., Zivy M.. (2011) MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics, 17 (11) 3572-7
- Virlouvet L., Jacquemot MP., Gerentes D., Corti H., Bouton S., Gilard F., Valot B., Trouverie J., Tcherkez G., Falque M., Damerval C., Rogowsky P., Perez P., Noctor G., Zivy M., Coursol S.. (2011) The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant physiology, 2 (157) 917-36


